In brief
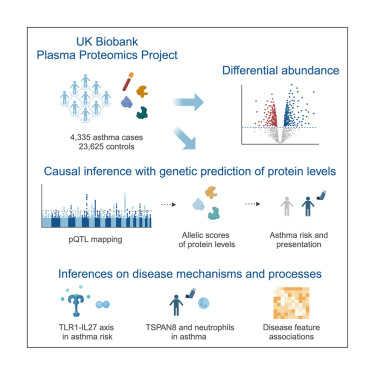
Plasma proteomics may capture key insights into disease mechanisms and reveal informative biomarkers. Donoghue et al. apply a causal inference framework to genetic and plasma proteomic data from the UK Biobank and demonstrate that genetically predicted protein levels are effective instruments for identifying putatively causal factors underlying both disease risk and heterogeneity.
Highlights
- Genetically predicted protein levels reveal putatively causal mechanisms in asthma
- Identification of candidate downstream biomarkers of asthma
- Causal protein candidates have differential associations with asthma endophenotypes
- Evidence for a TLR1-IL-27 axis in asthma risk and TSPAN8 in asthma neutrophil counts
Abstract
Hundreds of genetic associations for asthma have been identified, yet translating these findings into mechanistic insights remains challenging. We leveraged plasma proteomics from the UK Biobank Pharma Proteomics Project (UKB-PPP) to identify biomarkers and effectors of asthma risk or heterogeneity using genetic causal inference approaches. We identified 609 proteins associated with asthma status (269 proteins after controlling for body mass index [BMI] and smoking). Analysis of genetically predicted protein levels identified 70 proteins with putative causal roles in asthma risk, including known drug targets and proteins without prior genetic evidence in asthma (e.g., GCHFR, TDRKH, and CLEC7A). The genetic architecture of causally associated proteins provided evidence for a Toll-like receptor (TLR)1-interleukin (IL)-27 asthma axis. Lastly, we identified evidence of causal relationships between proteins and heterogeneous aspects of asthma biology, including between TSPAN8 and neutrophil counts. These findings illustrate that integrating biobank-scale genetics and plasma proteomics can provide a framework to identify therapeutic targets and mechanisms underlying disease risk and heterogeneity.